Authentic research experiences increase student retention and graduation rates. We know this “research effect” is even greater on underrepresented minorities in STEM. The Howard Hughes Medical Institute SEA-PHAGES program is an international undergraduate research program that gives students authentic research experience and has shown to increase student interest and retention in the sciences. It is designed as a two-semester course that begins with the discovery and isolation of novel bacteriophages (viruses that infect bacteria) in the first semester. Students continue to study these novel bacteriophages in the second semester by applying bioinformatics techniques to annotate the genomes (complete set of genetic material) of these bacteriophages.
This program provides an invaluable opportunity for students to experience authentic research in a structured manner and contribute to a large (and growing) body of scientific knowledge that has applications in biomedicine. Just this year, a cocktail of bacteriophages collected through the SEA-PHAGES program was used to successfully treat an antibiotic-resistant Mycobacterium infection in a 15-year-old patient with cystic fibrosis in London (. Our own students at CSN are joining this scientific endeavor, learning relevant skills, and beginning to see what a future in the sciences holds for them.
Conference Presentation: Harrington, D., Whiteby, O., Pochiro, S., Moriarty, M., Robertson, M., Stevens, J., Sanchez, A., Johnson, M., Theoret, J., Windsor, E., Yoon, E., Kimberley, K., and McKenna, C. The College of Southern Nevada’s Exploration into the DR and CS Clusters throughout the Las Vegas Valley. 11th SEA-PHAGES Symposium, Janelia Research Campus, Virginia. June 8, 2019.
For more information, please Dr. Chelsey Mckenna.
Published results:
Baumgartner BM, Bono KA, McIntosh DR, Vu AM, Adams CF, Benik BC, Chavez J, Gresky SJ, Sotelo A, Ray JI, Peister A, Kimberley KW, McKenna CC, Theoret JR, Yoon EJ, Windsor EJ. 2024. Genome sequences of actinobacteriophages JorRay, Blocker23, Nibbles, and OlgasClover. Microbiology Resource Announcements 0:e01256-23. https://doi.org/10.1128/mra.01256-23
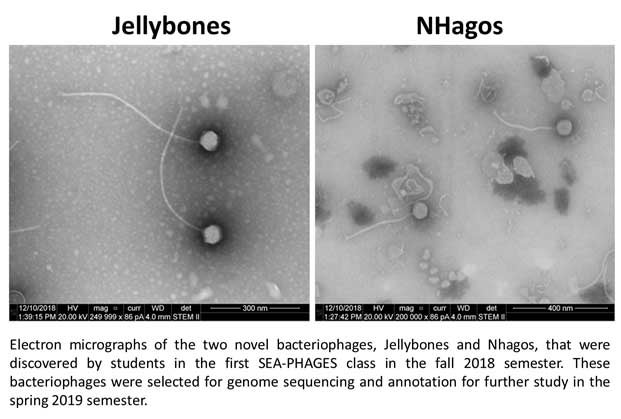
D'Andrew L. Harrington, Jennifer L. Stevens, Mia J. Johnson, Samantha J. Pochiro, Michelle M. Moriarty, Miriam E. Robertson, Andy Sanchez, Orin G. Whitby, Kendra W. Kimberley, Chelsey C. McKenna, James R. Theoret, Erin J. Windsor, Earl J. Yoon, 2020. Genome Sequences of Gordonia rubripertincta Bacteriophages Jellybones and NHagos. Microbiology Resource Announcements, 9(40): 1-3.
https://mra.asm.org/content/9/40/e00935-20
Harrington, DL, HR. Barten, EI. Audannio, FA. Bragg, KA. Garcia, KM. Niswonger, JR. MacAllister, AM. McCarroll, DM. O’Sullivan, SE. Torres, CD. Stanley, D Yalzadeh, KW. Kimberley, CC. McKenna, JR. Theoret, EJ. Yoon, EJ. Windsor, 2021. Genome Sequences of Gordonia Bacteriophages Jodelie19, BlingBling, and Burnsey. Microbiology Resource Announcements, 10 (5): e01280-20. https://mra.asm.org/content/10/5/e01280-20
Ethan Curran, Sabrina E. Callaway, Ramel R. Dumanlang, Anna V. Harshaw, Paula N. Palacio, Yuta Nakamura, Kendra W. Kimberley, James R. Theoret, Earl J. Yoon, Erin J. Windsor, Chelsey C. McKenna. 2022. Genome Sequences of Gordonia rubripertincta Bacteriophages AnarQue and Figliar. Microbiology Resource Announcements 11(1):e01085-21.https://journals.asm.org/doi/10.1128/mra.01085-21